Random forest in scikit-learn#
We illustrate the following regression method on a data set called “Hitters”, which includes 20 variables and 322 observations of major league baseball players. The goal is to predict a baseball player’s salary on the basis of various features associated with performance in the previous year. We don’t cover the topic of exploratory data analysis in this notebook.
Visit this documentation if you want to learn more about the data
Setup#
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from collections import OrderedDict
from sklearn.ensemble import RandomForestRegressor
from sklearn.metrics import mean_squared_error
from sklearn.inspection import permutation_importance
Data#
Import#
df = pd.read_csv("https://raw.githubusercontent.com/kirenz/datasets/master/Hitters.csv")
df
| AtBat | Hits | HmRun | Runs | RBI | Walks | Years | CAtBat | CHits | CHmRun | CRuns | CRBI | CWalks | League | Division | PutOuts | Assists | Errors | Salary | NewLeague | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 293 | 66 | 1 | 30 | 29 | 14 | 1 | 293 | 66 | 1 | 30 | 29 | 14 | A | E | 446 | 33 | 20 | NaN | A |
| 1 | 315 | 81 | 7 | 24 | 38 | 39 | 14 | 3449 | 835 | 69 | 321 | 414 | 375 | N | W | 632 | 43 | 10 | 475.0 | N |
| 2 | 479 | 130 | 18 | 66 | 72 | 76 | 3 | 1624 | 457 | 63 | 224 | 266 | 263 | A | W | 880 | 82 | 14 | 480.0 | A |
| 3 | 496 | 141 | 20 | 65 | 78 | 37 | 11 | 5628 | 1575 | 225 | 828 | 838 | 354 | N | E | 200 | 11 | 3 | 500.0 | N |
| 4 | 321 | 87 | 10 | 39 | 42 | 30 | 2 | 396 | 101 | 12 | 48 | 46 | 33 | N | E | 805 | 40 | 4 | 91.5 | N |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 317 | 497 | 127 | 7 | 65 | 48 | 37 | 5 | 2703 | 806 | 32 | 379 | 311 | 138 | N | E | 325 | 9 | 3 | 700.0 | N |
| 318 | 492 | 136 | 5 | 76 | 50 | 94 | 12 | 5511 | 1511 | 39 | 897 | 451 | 875 | A | E | 313 | 381 | 20 | 875.0 | A |
| 319 | 475 | 126 | 3 | 61 | 43 | 52 | 6 | 1700 | 433 | 7 | 217 | 93 | 146 | A | W | 37 | 113 | 7 | 385.0 | A |
| 320 | 573 | 144 | 9 | 85 | 60 | 78 | 8 | 3198 | 857 | 97 | 470 | 420 | 332 | A | E | 1314 | 131 | 12 | 960.0 | A |
| 321 | 631 | 170 | 9 | 77 | 44 | 31 | 11 | 4908 | 1457 | 30 | 775 | 357 | 249 | A | W | 408 | 4 | 3 | 1000.0 | A |
322 rows × 20 columns
df.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 322 entries, 0 to 321
Data columns (total 20 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 AtBat 322 non-null int64
1 Hits 322 non-null int64
2 HmRun 322 non-null int64
3 Runs 322 non-null int64
4 RBI 322 non-null int64
5 Walks 322 non-null int64
6 Years 322 non-null int64
7 CAtBat 322 non-null int64
8 CHits 322 non-null int64
9 CHmRun 322 non-null int64
10 CRuns 322 non-null int64
11 CRBI 322 non-null int64
12 CWalks 322 non-null int64
13 League 322 non-null object
14 Division 322 non-null object
15 PutOuts 322 non-null int64
16 Assists 322 non-null int64
17 Errors 322 non-null int64
18 Salary 263 non-null float64
19 NewLeague 322 non-null object
dtypes: float64(1), int64(16), object(3)
memory usage: 50.4+ KB
Missing values#
Note that the salary is missing for some of the players:
print(df.isnull().sum())
AtBat 0
Hits 0
HmRun 0
Runs 0
RBI 0
Walks 0
Years 0
CAtBat 0
CHits 0
CHmRun 0
CRuns 0
CRBI 0
CWalks 0
League 0
Division 0
PutOuts 0
Assists 0
Errors 0
Salary 59
NewLeague 0
dtype: int64
We simply drop the missing cases:
# drop missing cases
df = df.dropna()
Create label and features#
Since we will use algorithms from scikit learn, we need to encode our categorical features as one-hot numeric features (dummy variables):
dummies = pd.get_dummies(df[['League', 'Division','NewLeague']])
dummies.info()
<class 'pandas.core.frame.DataFrame'>
Int64Index: 263 entries, 1 to 321
Data columns (total 6 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 League_A 263 non-null uint8
1 League_N 263 non-null uint8
2 Division_E 263 non-null uint8
3 Division_W 263 non-null uint8
4 NewLeague_A 263 non-null uint8
5 NewLeague_N 263 non-null uint8
dtypes: uint8(6)
memory usage: 3.6 KB
print(dummies.head())
League_A League_N Division_E Division_W NewLeague_A NewLeague_N
1 0 1 0 1 0 1
2 1 0 0 1 1 0
3 0 1 1 0 0 1
4 0 1 1 0 0 1
5 1 0 0 1 1 0
Next, we create our label y:
y = df[['Salary']]
We drop the column with the outcome variable (Salary), and categorical columns for which we already created dummy variables:
X_numerical = df.drop(['Salary', 'League', 'Division', 'NewLeague'], axis=1).astype('float64')
Make a list of all numerical features (we need them later):
list_numerical = X_numerical.columns
list_numerical
Index(['AtBat', 'Hits', 'HmRun', 'Runs', 'RBI', 'Walks', 'Years', 'CAtBat',
'CHits', 'CHmRun', 'CRuns', 'CRBI', 'CWalks', 'PutOuts', 'Assists',
'Errors'],
dtype='object')
# Create all features
X = pd.concat([X_numerical, dummies[['League_N', 'Division_W', 'NewLeague_N']]], axis=1)
X.info()
<class 'pandas.core.frame.DataFrame'>
Int64Index: 263 entries, 1 to 321
Data columns (total 19 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 AtBat 263 non-null float64
1 Hits 263 non-null float64
2 HmRun 263 non-null float64
3 Runs 263 non-null float64
4 RBI 263 non-null float64
5 Walks 263 non-null float64
6 Years 263 non-null float64
7 CAtBat 263 non-null float64
8 CHits 263 non-null float64
9 CHmRun 263 non-null float64
10 CRuns 263 non-null float64
11 CRBI 263 non-null float64
12 CWalks 263 non-null float64
13 PutOuts 263 non-null float64
14 Assists 263 non-null float64
15 Errors 263 non-null float64
16 League_N 263 non-null uint8
17 Division_W 263 non-null uint8
18 NewLeague_N 263 non-null uint8
dtypes: float64(16), uint8(3)
memory usage: 35.7 KB
# Create a list of feature names
feature_names = X.columns
Split data#
Split the data set into train and test set with the first 70% of the data for training and the remaining 30% for testing.
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=10)
X_train.head()
| AtBat | Hits | HmRun | Runs | RBI | Walks | Years | CAtBat | CHits | CHmRun | CRuns | CRBI | CWalks | PutOuts | Assists | Errors | League_N | Division_W | NewLeague_N | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 260 | 496.0 | 119.0 | 8.0 | 57.0 | 33.0 | 21.0 | 7.0 | 3358.0 | 882.0 | 36.0 | 365.0 | 280.0 | 165.0 | 155.0 | 371.0 | 29.0 | 1 | 1 | 1 |
| 92 | 317.0 | 78.0 | 7.0 | 35.0 | 35.0 | 32.0 | 1.0 | 317.0 | 78.0 | 7.0 | 35.0 | 35.0 | 32.0 | 45.0 | 122.0 | 26.0 | 0 | 0 | 0 |
| 137 | 343.0 | 103.0 | 6.0 | 48.0 | 36.0 | 40.0 | 15.0 | 4338.0 | 1193.0 | 70.0 | 581.0 | 421.0 | 325.0 | 211.0 | 56.0 | 13.0 | 0 | 0 | 0 |
| 90 | 314.0 | 83.0 | 13.0 | 39.0 | 46.0 | 16.0 | 5.0 | 1457.0 | 405.0 | 28.0 | 156.0 | 159.0 | 76.0 | 533.0 | 40.0 | 4.0 | 0 | 1 | 0 |
| 100 | 495.0 | 151.0 | 17.0 | 61.0 | 84.0 | 78.0 | 10.0 | 5624.0 | 1679.0 | 275.0 | 884.0 | 1015.0 | 709.0 | 1045.0 | 88.0 | 13.0 | 0 | 0 | 0 |
y_train
| Salary | |
|---|---|
| 260 | 875.0 |
| 92 | 70.0 |
| 137 | 430.0 |
| 90 | 431.5 |
| 100 | 2460.0 |
| ... | ... |
| 274 | 200.0 |
| 196 | 587.5 |
| 159 | 200.0 |
| 17 | 175.0 |
| 162 | 75.0 |
184 rows × 1 columns
Data standardization#
Some of our models perform best when all numerical features are centered around 0 and have variance in the same order (like Lasso, Ridge or GAMs).
To avoid data leakage, the standardization of numerical features should always be performed after data splitting and only from training data.
Furthermore, we obtain all necessary statistics for our features (mean and standard deviation) from training data and also use them on test data. Note that we don’t standardize our dummy variables (which only have values of 0 or 1).
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler().fit(X_train[list_numerical])
X_train[list_numerical] = scaler.transform(X_train[list_numerical])
X_test[list_numerical] = scaler.transform(X_test[list_numerical])
Make contiguous flattened arrays (for our scikit-learn model):
y_train = np.ravel(y_train)
y_test = np.ravel(y_test)
Model#
Define hyperparameters:
params = {
"n_estimators": 500,
"max_depth": 4,
"min_samples_split": 5,
"warm_start":True,
"oob_score":True,
"random_state": 42,
}
Build and fit model
reg =RandomForestRegressor(**params)
reg.fit(X_train, y_train)
RandomForestRegressor(max_depth=4, min_samples_split=5, n_estimators=500,
oob_score=True, random_state=42, warm_start=True)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
RandomForestRegressor(max_depth=4, min_samples_split=5, n_estimators=500,
oob_score=True, random_state=42, warm_start=True)Make predictions
y_pred = reg.predict(X_test)
Evaluate model with RMSE
mean_squared_error(y_test, y_pred, squared=False)
296.37036964432764
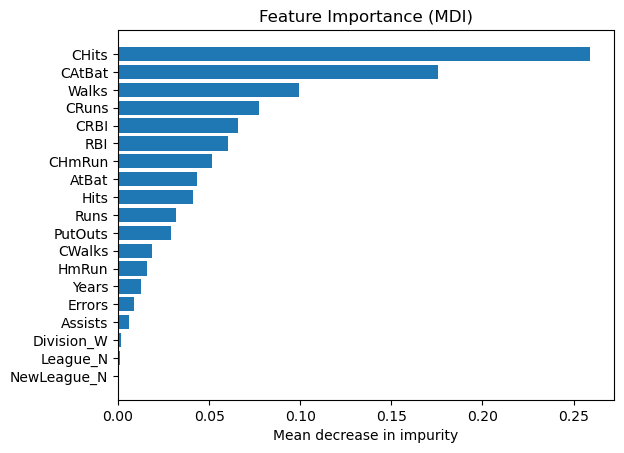
Feature importance#
Next, we take a look at the tree based feature importance and the permutation importance.
Mean decrease in impurity (MDI)#
Mean decrease in impurity (MDI) is a measure of feature importance for decision tree models.
Note
Visit this notebook to learn more about MDI
Feature importances are provided by the fitted attribute
feature_importances_
# obtain feature importance
feature_importance = reg.feature_importances_
# sort features according to importance
sorted_idx = np.argsort(feature_importance)
pos = np.arange(sorted_idx.shape[0])
# plot feature importances
plt.barh(pos, feature_importance[sorted_idx], align="center")
plt.yticks(pos, np.array(feature_names)[sorted_idx])
plt.title("Feature Importance (MDI)")
plt.xlabel("Mean decrease in impurity");
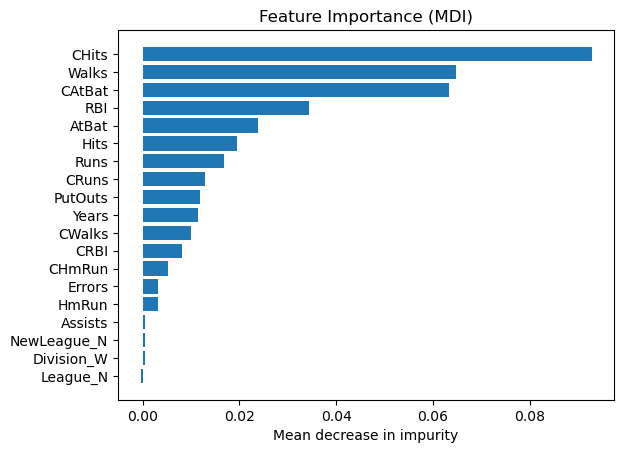
Permutation feature importance#
The permutation feature importance is defined to be the decrease in a model score when a single feature value is randomly shuffled.
Note
Visit this notebook to learn more about permutation feature importance.
result = permutation_importance(
reg, X_test, y_test, n_repeats=10, random_state=42, n_jobs=2
)
tree_importances = pd.Series(result.importances_mean, index=feature_names)
# sort features according to importance
sorted_idx = np.argsort(tree_importances)
pos = np.arange(sorted_idx.shape[0])
# plot feature importances
plt.barh(pos, tree_importances[sorted_idx], align="center")
plt.yticks(pos, np.array(feature_names)[sorted_idx])
plt.title("Feature Importance (MDI)")
plt.xlabel("Mean decrease in impurity");
Same data plotted as boxplot:
plt.boxplot(
result.importances[sorted_idx].T,
vert=False,
labels=np.array(feature_names)[sorted_idx],
)
plt.title("Permutation Importance (test set)")
Text(0.5, 1.0, 'Permutation Importance (test set)')
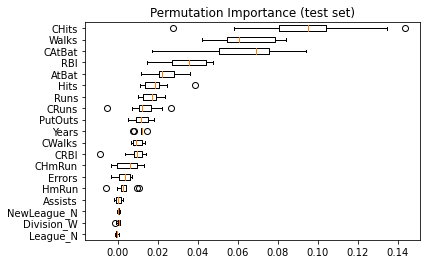
We observe that the same features are detected as most important using both methods (e.g.,
CAtBat,CRBI,CHits,Walks,Years). Although the relative importances vary (especially for featureYears).

